We are excited to see SPLASH highlighted at RECOMB 2024 highlight session!
Hello To Our New Members
Welcome to our new members Khoa Hoang, Conor Messer, and Daniel Cotte, who will be joining the lab this spring!
Julie Wang joins the lab!
Welcome Julie!
Tavor and Elisabeth graduate!
Congratulations to Tavor and Elisabeth!
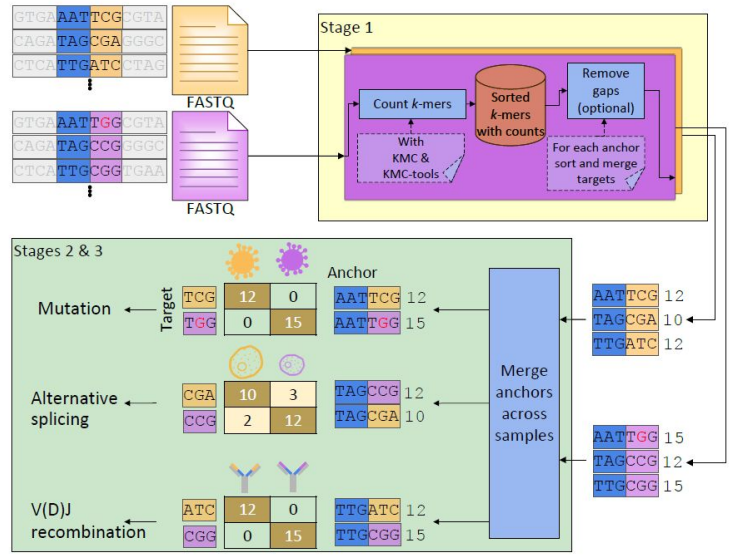
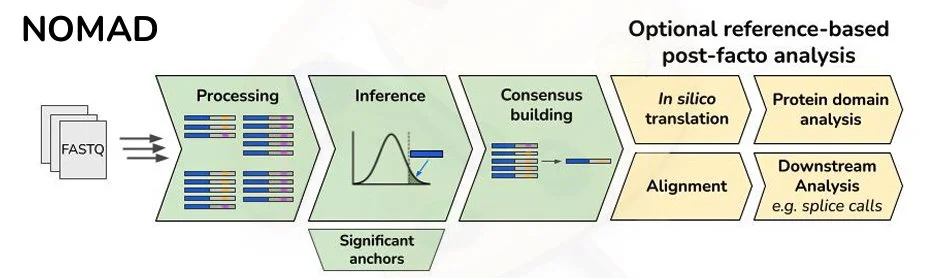
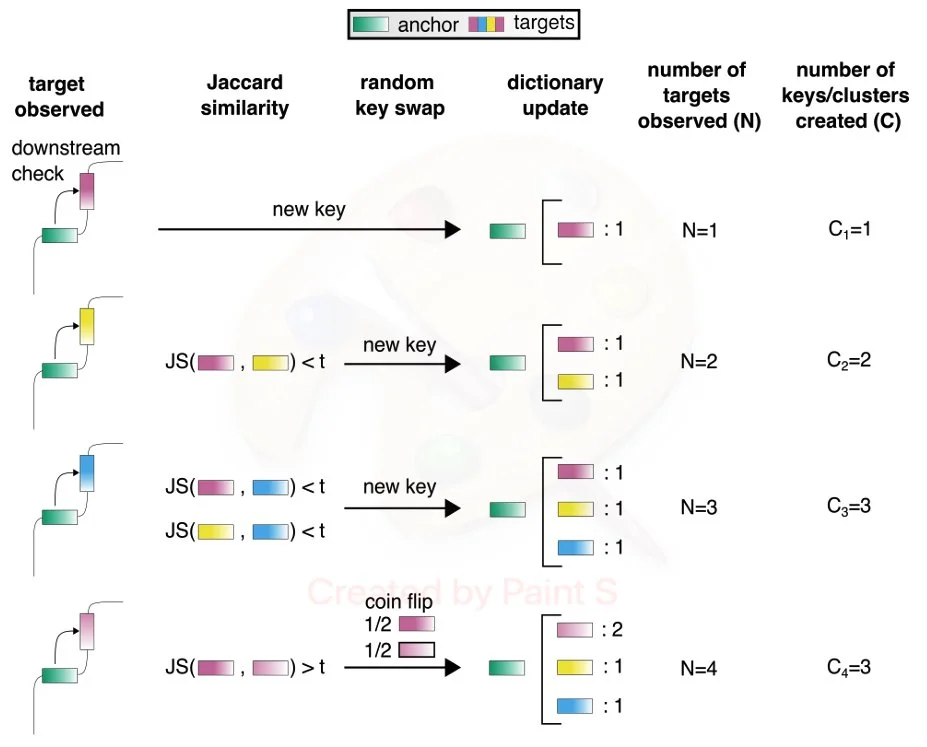
New Preprint on Ultra-Efficient, Scalable and Unsupervised Discovery with SPLASH2
Congrats to Marek, Roozbeh, and Tavor on publishing SPLASH2! very excited to see what SPLASH2 can discover using its statistical power and computational speed.
best wishes for our recent alumni and Hello to new members
We end 2022 by saying goodbye to Kaitlin, Rob, and Jordi and welcoming George, Elinor, and Punit! Best of luck to our previous members and excited to have our new members in the lab!
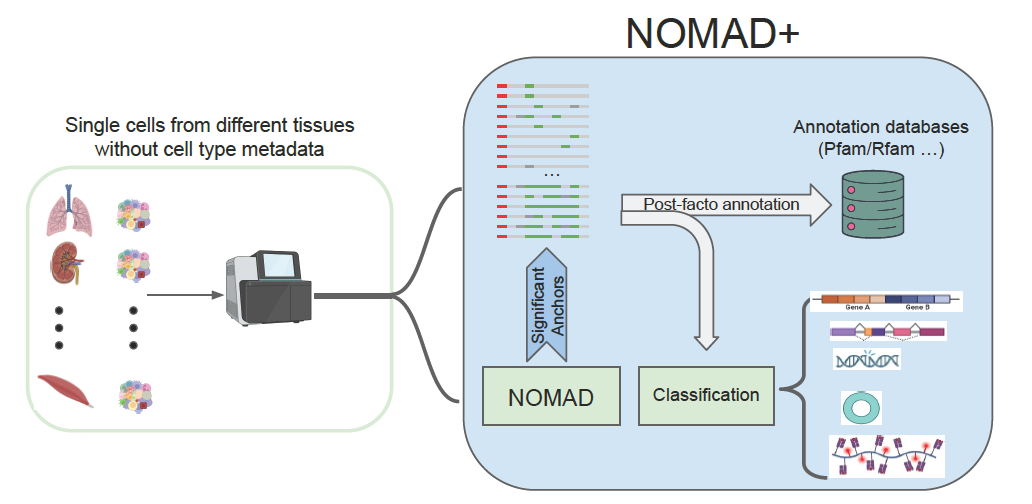
New preprint on unsupervised reference-free inference of single-cell transcriptomic complexity
Congrats to the everyone involved in this massive project on unbiased reference-free detection of RNA regulation and diversification in single cells via NOMAD+.
SPLASH Preprint Now Online
Excited to share a new preprint on SPLASH, a statistical reference-free approach for direct inference on raw reads. Great job Kaitlin and Tavor!
DIVE Preprint Published On BioRxiv
Congratulations to Jordi and Peter for “DIVE“ing into diversity-generating and mobile genetic elements!
Goodbye to Julia Olivieri
Congratulations to Julia Olivieri for graduating with her PhD!
Welcome Tavor!
We are very excited to have Tavor Baharav join the Salzman lab! Tavor is currently working on designing efficient data-driven algorithms that adapt to problem structure by leveraging techniques from multi-armed bandits.
Tabula Sapiens paper out now in Science
Congratulations to Roozbeh and Julia for their contributions to the Tabula Sapiens paper!
Congratulations to Julia Olivieri on her successful thesis defense!
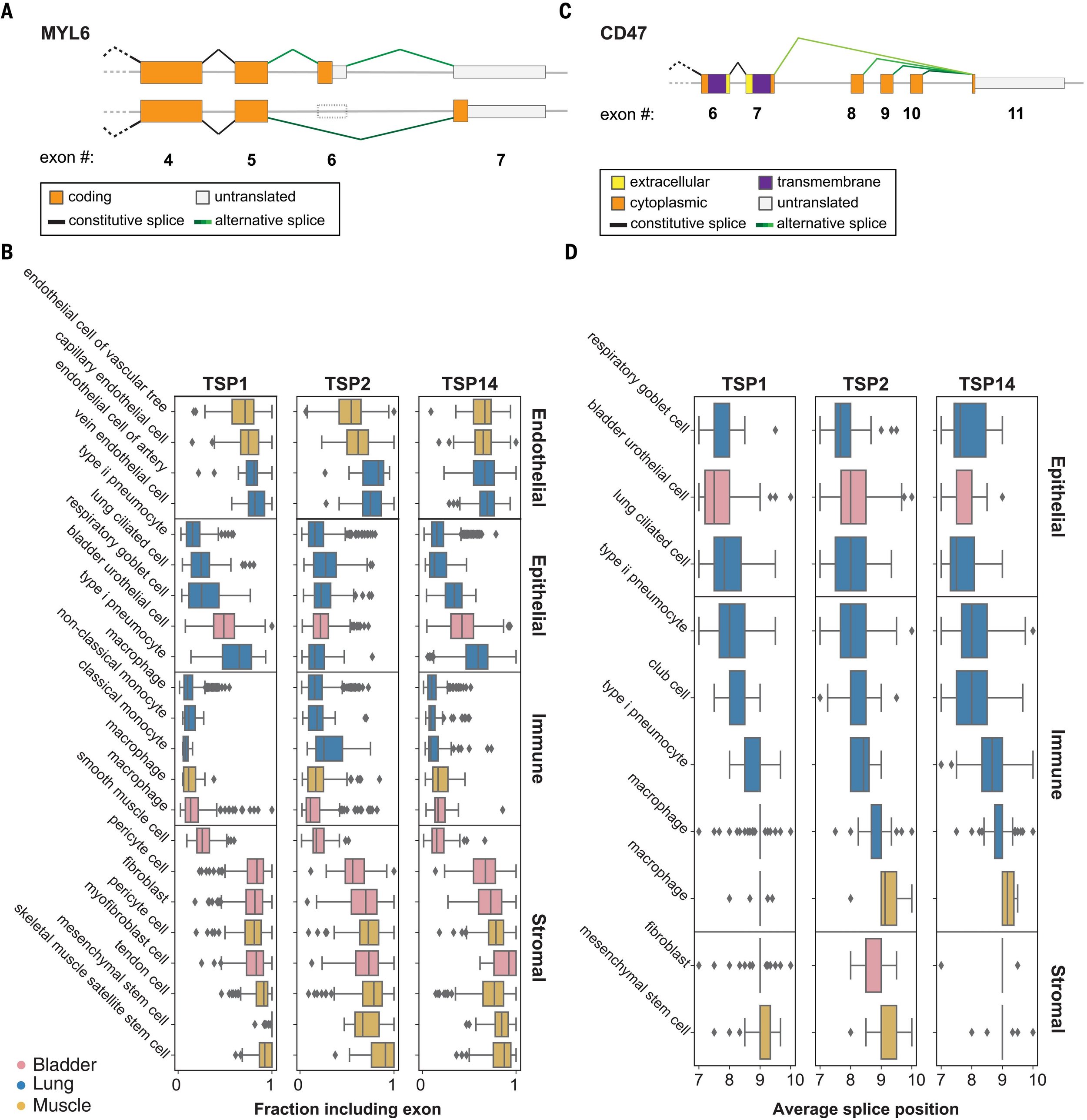
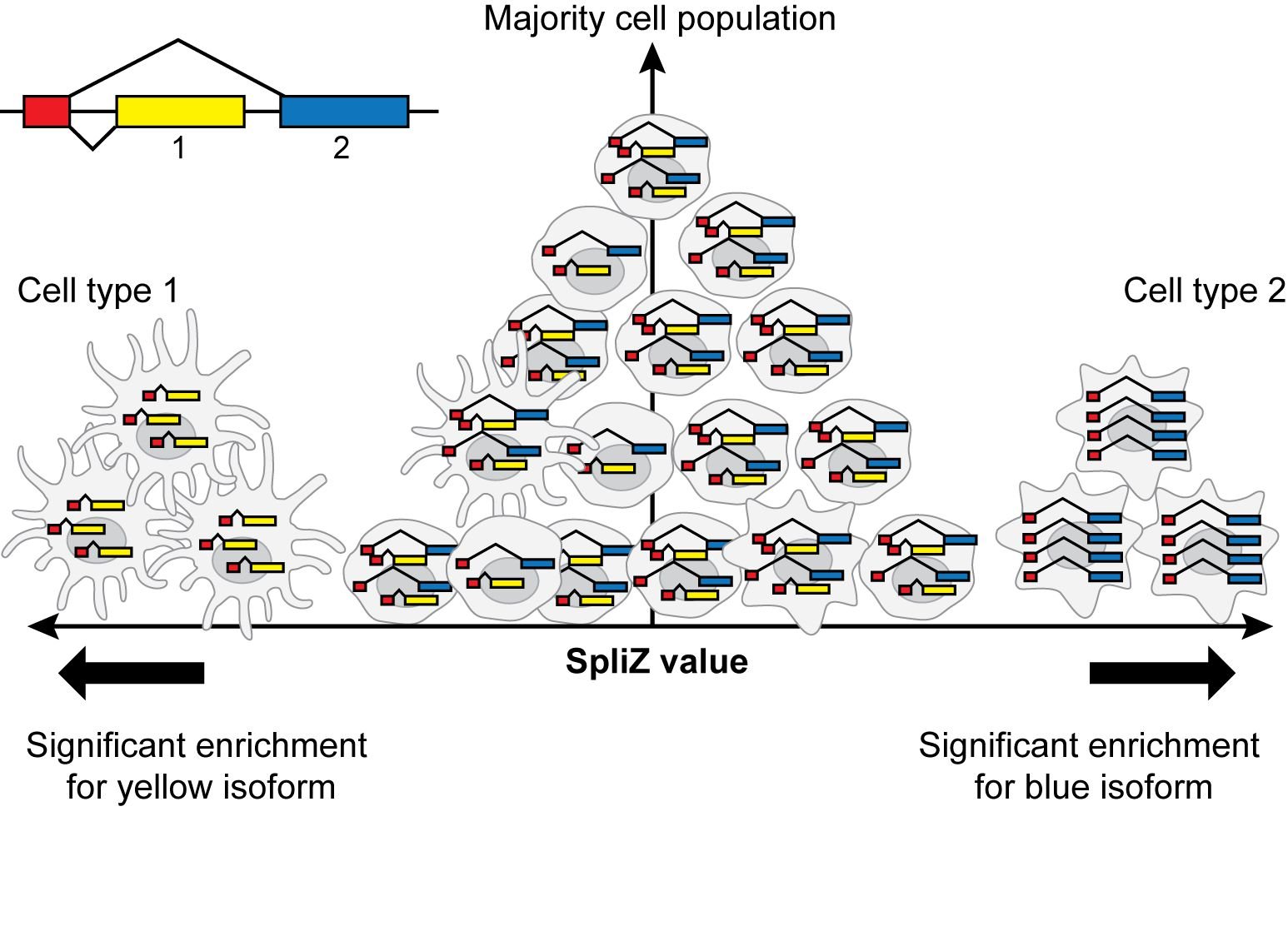
Paper introducing the SpliZ method accepted at Nature Methods
The manuscript “The SpliZ generalizes “Percent Spliced In” to reveal regulated splicing at single-cell resolution” led by Julia Olivieri and Roozbeh Dehghannasiri has been accepted for publication at Nature Methods.
Abstract: Detecting single-cell-regulated splicing from droplet-based technologies has been viewed as out of reach. Here, we introduce the Splicing Z Score (SpliZ), an annotation-free statistical method to detect regulated splicing in single-cell RNA-seq. We applied the SpliZ to human lung cells, discovering hundreds of genes with cell-type-specific splicing patterns including ones with potential implications for basic and translational biology.
Visiting student Marieke joins the lab!
Marieke Vromman will be visiting from Ghent University through June 2022. She’ll be working on expanding the analysis of splicing in single cells. Welcome Marieke!
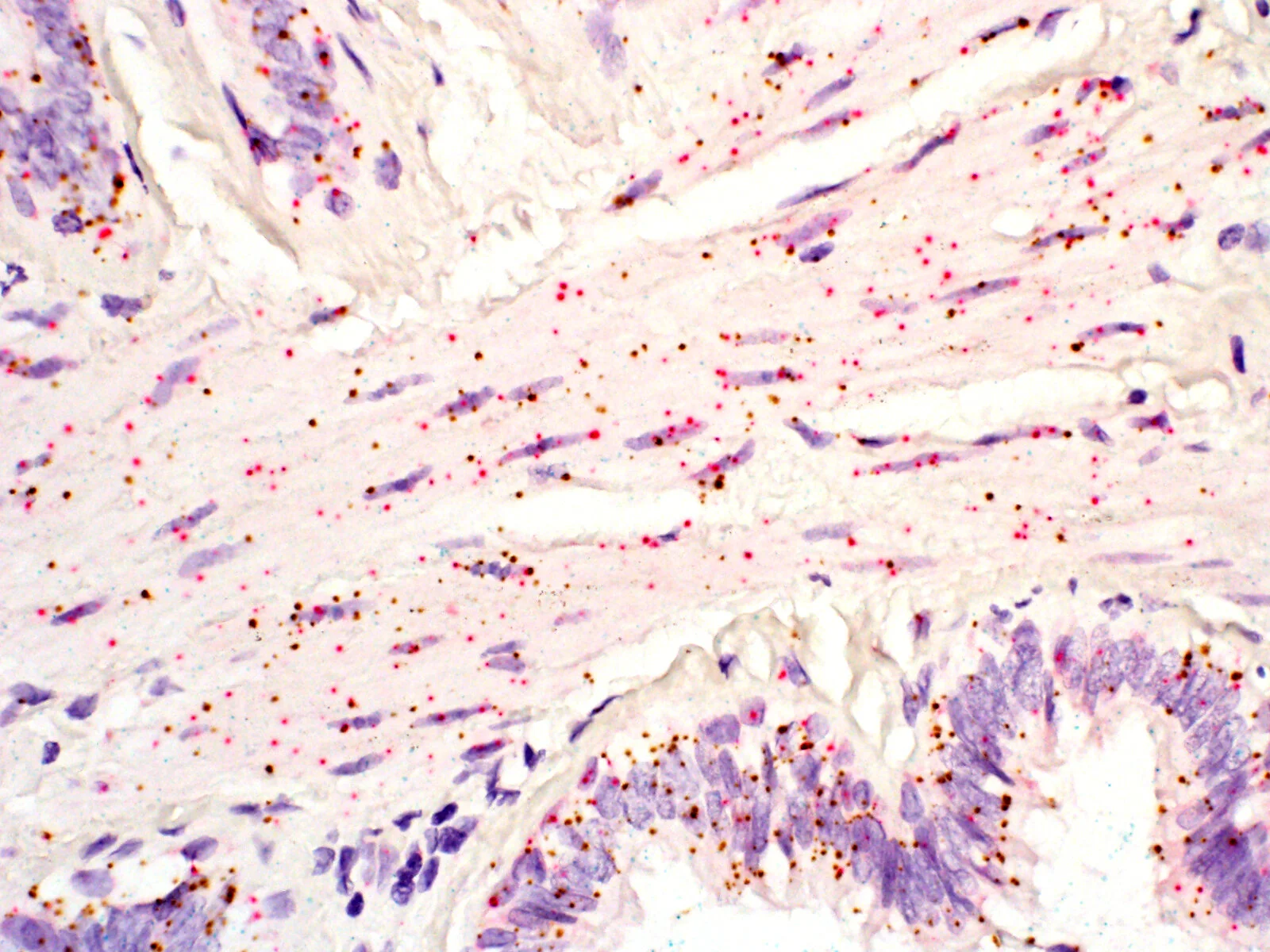
Paper analyzing splicing in the Tabula Sapiens dataset accepted at eLife
The manuscript “RNA splicing programs define tissue compartments and cell types at single cell resolution” led by Julia Olivieri and Roozbeh Dehghannasiri has been accepted for publication at eLife (https://elifesciences.org/articles/70692).
Abstract: The extent splicing is regulated at single-cell resolution has remained controversial due to both available data and methods to interpret it. We apply the SpliZ, a new statistical approach, to detect cell-type-specific splicing in >110K cells from 12 human tissues. Using 10x data for discovery, 9.1% of genes with computable SpliZ scores are cell-type-specifically spliced, including ubiquitously expressed genes MYL6 and RPS24. These results are validated with RNA FISH, single-cell PCR, and Smart-seq2. SpliZ analysis reveals 170 genes with regulated splicing during human spermatogenesis, including examples conserved in mouse and mouse lemur. The SpliZ allows model-based identification of subpopulations indistinguishable based on gene expression, illustrated by subpopulation-specific splicing of classical monocytes involving an ultraconserved exon in SAT1. Together, this analysis of differential splicing across multiple organs establishes that splicing is regulated cell-type-specifically.
Vicky Cepeda joins the lab full time
Welcome to Vicky, who will be joining the lab as a full-time postdoc! Vicky will be working on analyzing expression of RNA following viral infection.
Luis Chumpitaz Rotates in!
We are happy to announce that Luis Chumpitaz will rotate with our lab for the upcoming quarter!
Jordi Abante awarded a CEHG Fellowship!
Congratulations to Jordi Abante, who was awarded a postdoctoral fellowship from Stanford’s Center for Computational, Evolutionary and Human Genomics (CEHG)! More information about the fellowship can be found here: https://cehg.stanford.edu/fellowship-program
Lieberman Fellowship Awarded
Congratulations to Julia Olivieri, who received a Lieberman Fellowship from Stanford for 2021-2022! More details can be found here: https://vpge.stanford.edu/fellowships-funding/gerald-j-lieberman-fellowship/details