Selected Publications
Ultra-efficient, unified discovery from microbial sequencing with SPLASH and precise statistical assembly [URL]
G. Henderson, A. Gudys, T. Baharav, P. Sundaramurthy, M. Kokot, P. L. Wang, S. Deorowicz, A. Carey, J. Salzman.
bioRxiv. (2024)
SPLASH2 provides ultra-efficient, scalable, and unsupervised discovery on raw sequencing reads [URL]
M. Kokot*, R. Dehghannasiri*, T.Z. Baharav, J. Salzman†, S. Deorowicz†
bioRxiv (2023)
Unsupervised reference-free inference reveals unrecognized regulated transcriptomic complexity in human single cells [URL]
R. Dehghannasiri*, G Henderson*, R. Bierman, K. Chaung, T. Baharav, P. Wang, J. Salzman
bioRxiv (2023)
RNA splicing programs define tissue compartments and cell types at single cell resolution
J. Olivieri*, R. Dehghannasiri*, P. Wang, S. Jang, A. de Morree, S. Tan, J. Ming, A. Wu, Tabula Sapiens Consortium, S. Quake, M. Krasnow, J. Salzman (2021). eLife. https://elifesciences.org/articles/70692
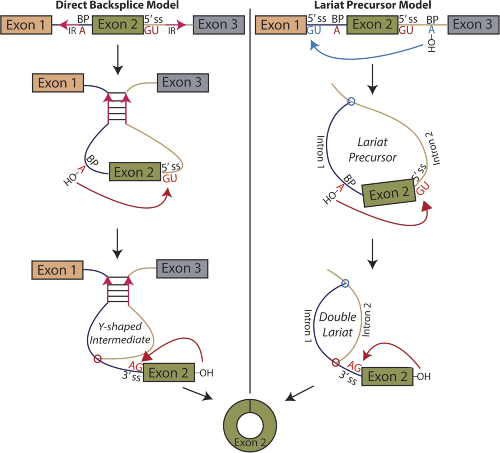
Circular RNA biogenesis can proceed through an exon-containing lariat precursor
S.P. Barrett, P.L. Wang, J. Salzman (2015) eLife. http://elifesciences.org/content/elife/4/e07540.full.pdf
Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during fetal development
L. Szabo, R. Morey, N.J. Palpant, P.L. Wang, N. Afari, C. Jiang, M.M. Parast, C.E. Murry, L.C. Laurent, J. Salzman (2015). Genome Biology. https://pubmed.ncbi.nlm.nih.gov/26076956/
All Publications
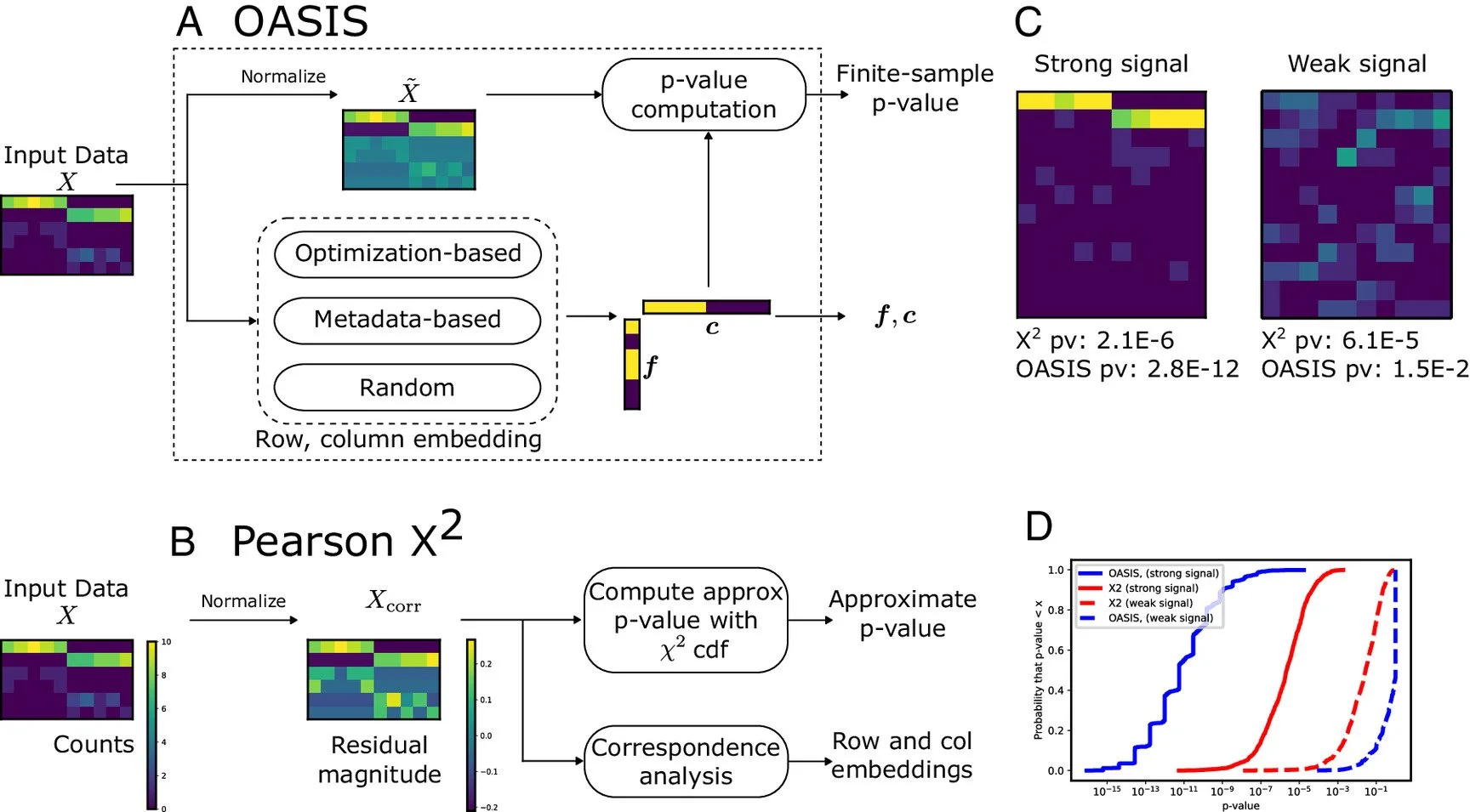
Baharav, T. Z., Tse, D., & Salzman, J. (2024). “OASIS: An Interpretable, Finite-Sample Valid Alternative to Pearson’sx2for Scientific Discovery”. PNAS, https://doi.org/10.1073/pnas.2304671121
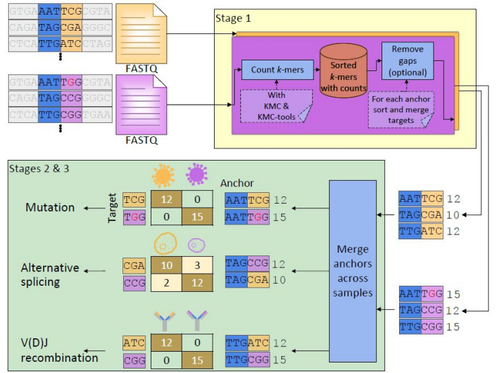
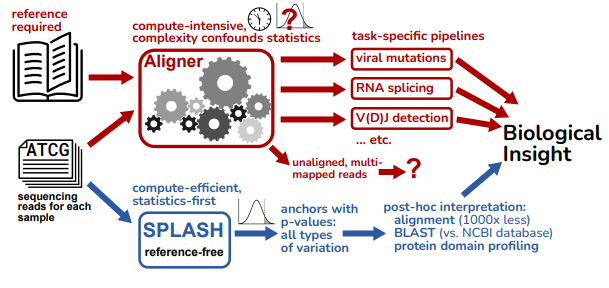
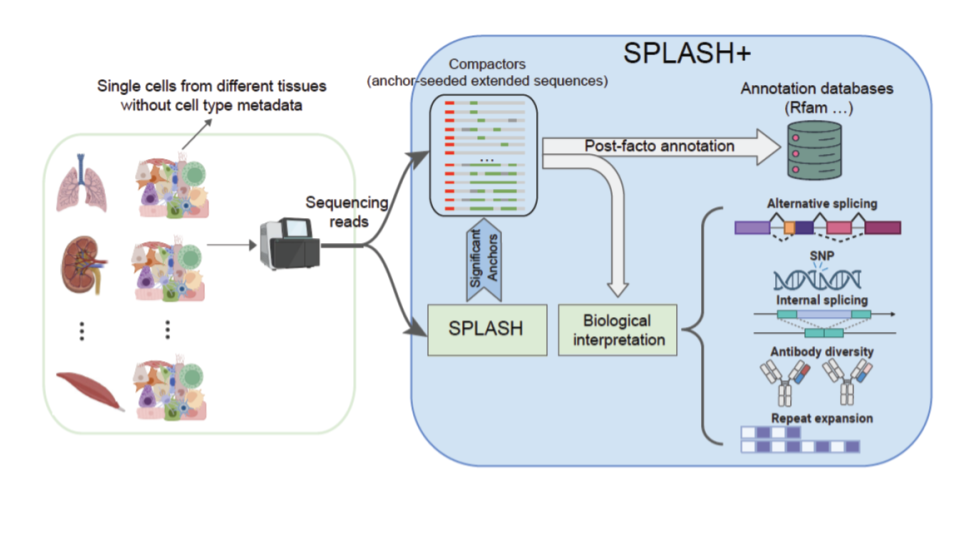
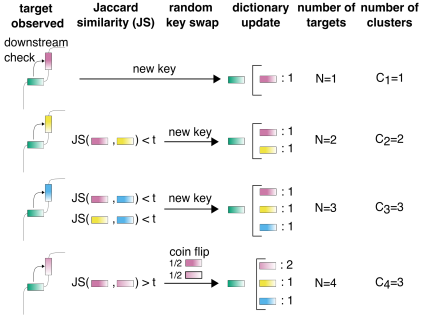
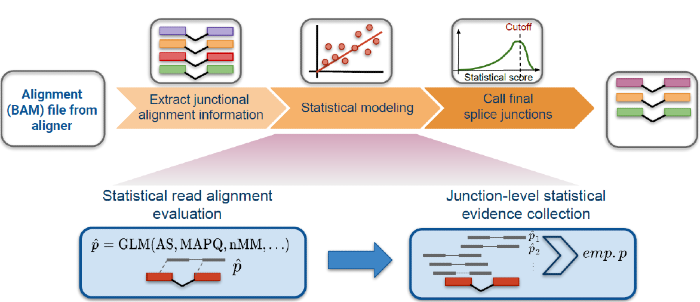
K. Chaung*, T. Baharav*, G. Henderson, I. Zheludev, P. L. Wang, J. Salzman (2023). “SPLASH: a statistical, reference-free genomic algorithm unifies biological discovery”. Cell. https://doi.org/10.1016/j.cell.2023.10.028
G. Henderson*, A. Gudys*, T. Baharav, P. Sundaramurthy, M. Kokot, P. L. Wang, S. Deorowicz, A. Carey, J. Salzman (2024). Ultra-efficient, unified discovery from microbial sequencing with SPLASH and precise statistical assembly. bioRxiv. https://www.biorxiv.org/content/10.1101/2024.01.18.576133v1
M. Kokot*, R. Dehghannasiri*, T.Z. Baharav, J. Salzman†, S. Deorowicz†, (2023). SPLASH2 provides ultra-efficient, scalable, and unsupervised discovery on raw sequencing reads. bioRxiv. https://doi.org/10.1101/2023.03.17.533189
R. Dehghannasiri*, G Henderson*, R. Bierman, K. Chaung, T. Baharav, P. Wang, J. Salzman (2023). “Unsupervised reference-free inference reveals unrecognized regulated transcriptomic complexity in human single cells” bioRxiv. https://doi.org/10.1101/2022.12.06.519414.
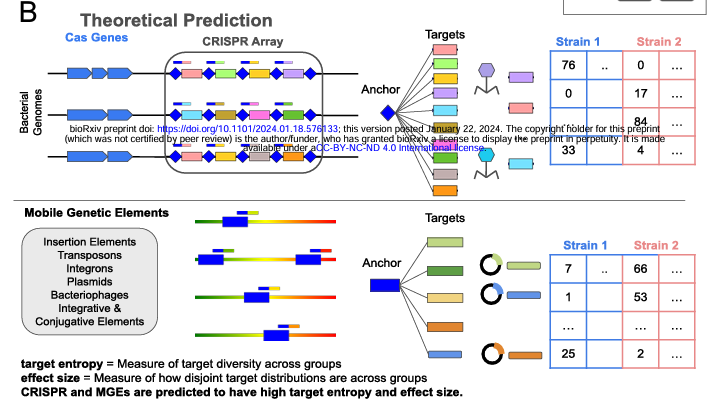
J. Abante, P. Wang, J. Salzman (2023). “DIVE: a reference-free statistical approach to diversity-generating & mobile genetic element discovery.” Genome Biology. https://doi.org/10.1186/s13059-023-03038-0
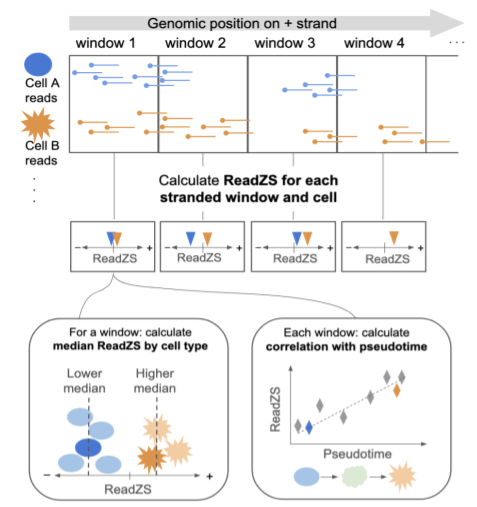
E. Meyer*, K. Chaung*, R. Dehghannasiri, J. Salzman (2022). “ReadZS detects cell type-specific and developmentally regulated RNA processing programs in single-cell RNA-seq.” Genome Biology. https://doi.org/10.1186/s13059-022-02795-8
The Tabula Sapiens Consortium (2022). “The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans.” Science. https://doi.org/10.1126/science.abl4896
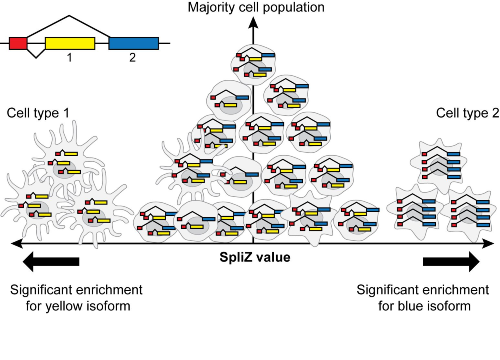
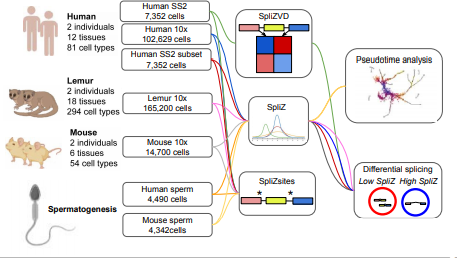
J. Olivieri*, R. Dehghannasiri*, J. Salzman (2022). "The SpliZ generalizes ‘Percent Spliced In’ to reveal regulated splicing at single-cell resolution." Nature Methods. https://www.nature.com/articles/s41592-022-01400-x
J. Olivieri*, R. Dehghannasiri*, P. Wang, S. Jang, A. de Morree, S. Tan, J. Ming, A. Wu, Tabula Sapiens Consortium, S. Quake, M. Krasnow, J. Salzman (2021). “RNA splicing programs define tissue compartments and cell types at single cell resolution.” eLife. https://elifesciences.org/articles/70692
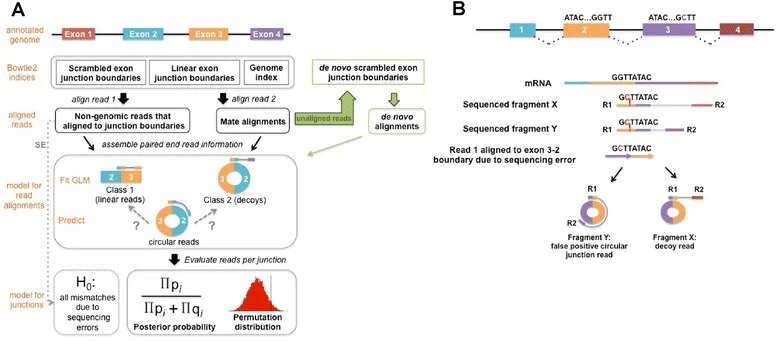
R. Dehghannasiri*, J. Olivieri*, A. Damljanovic, J. Salzman (2021). "Specific splice junction detection in single cells with SICILIAN", Genome Biology. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-021-02434-8
C. Horn, J. Salzman (2019). “Molecular sampling at logarithmic rates for next-generation sequencing”, PLoS Computational Biology. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007537
R. Dehghannasiri, D. Freeman, M. Jordanski, G. Hsieh, A. Damljanovic, E. Lehnert, J. Salzman (2019). “Improved detection of gene fusions by applying statistical methods reveals new oncogenic RNA cancer drivers”, Proceedings of National Academy of Sciences. https://www.pnas.org/doi/10.1073/pnas.1900391116
R. Dehghannasiri, L. Szabo, J. Salzman (2019). “Ambiguous splice sites distinguish circRNA and linear splicing in the human genome”, Bioinformatics. https://academic.oup.com/bioinformatics/article/35/8/1263/5091181
Barrett SP*, Parker KR*, Horn C, Mata M, Salzman J (2017). “ciRS-7 exonic sequence is embedded in a long non-coding RNA locus”, PLOS Genetics.
http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007114
G Hsieh, R Bierman, L Szabo, AG Lee, DE Freeman, N Watson, EA Sweet-Cordero, J Salzman (2017). “Statistical algorithms improve accuracy of gene fusion detection”, Nucleic Acids Research.
https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkx453
L Szabo, J Salzman (2016). “Detecting Circular RNAs: bioinformatic and experimental challenges”, Nature Reviews Genetics.
http://www.nature.com/nrg/journal/v17/n11/full/nrg.2016.114.html
S.P. Barrett, J Salzman (2016). Circular RNAs: analysis, expressions and potential functions. DOI 10.1242/dev.128074
http://dev.biologists.org/content/143/11/1838.abstract PDF
J Salzman (2016). “Circular RNA Expression: Its Potential Regulation and Function”, Trends in Genetics. http://www.sciencedirect.com/science/article/pii/S0168952516000329
S.P. Barrett, P.L. Wang, J. Salzman (2015). “Circular RNA biogenesis can proceed through an exon-containing lariat precursor”, eLife.
http://elifesciences.org/content/elife/4/e07540.full.pdf
L. Szabo, R. Morey, N.J. Palpant, P.L. Wang, N. Afari, C. Jiang, M.M. Parast, C.E. Murry, L.C. Laurent, J. Salzman (2015). “Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during fetal development”, Genome Biology.
https://pubmed.ncbi.nlm.nih.gov/26076956/
H. Jiang, J. Salzman (2014). “A penalized likelihood approach for robust estimation of isoform expression”, Statistics and Its Interface.
http://intlpress.com/site/pub/files/_fulltext/journals/sii/2015/0008/0004/SII-2015-0008-0004-a003.pdf
P.L. Wang, Y. Bao, M.C. Yee, S.P. Barrett, G.J. Hogan, M.N. Olsen, J.R. Dinneny, P.O. Brown, J. Salzman (2014). “Circular RNA Is Expressed across the Eukaryotic Tree of Life”, PLoS ONE.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0090859
J. Salzman, R.E. Chen, M.N. Olsen, P.L. Wang, P.O. Brown (2013). “Cell type specific features of circular RNA expression”, PLOS Genetics. http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1003777
B. Rajaratnam and J. Salzman (2013). “Best Permutation Analysis”, Journal of Multivariate Analysis.
https://www.sciencedirect.com/science/article/pii/S0047259X13000286
H. Jiang, J. Salzman (2012). “Statistical properties of an early stopping rule for resampling-based multiple testing”, Biometrika.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3629857/pdf/ass051.pdf
J. Salzman, C. Gawad, P. L. Wang, N. Lacayo and P. O. Brown (2012). “Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types”, PLOS One.
Faculty of 1000; features in Nature, Biotechniques and other scientific press venues.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0030733
J. Salzman, R. J. Marinelli, P. L. Wang, A. Greene, J. Nielsen, B. Nelsen, C. Dresher and P. O. Brown (2011). “ESRRA-C11orf20 is a Recurrent Gene Fusion in Serous Ovarian Carcinoma”, PLOS Biology.
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1001156
J. Salzman, H. Jiang, and W. Wong (2011). “Statistical Modeling of RNA-Seq Data”, Statistical Science.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3846358/pdf/nihms498524.pdf
D. T. Pride, C. Sun, J. Salzman, N. Rao, P. Loomer, G. C. Armitage, J. Banfield, and D. A. Relman (2010). “Analysis of Streptococcal CRISPRs from Human Saliva Reveals Substantial Sequence Diversity Within and Between Subjects Over Time”, Genome Research.
http://genome.cshlp.org/content/21/1/126.full.pdf
N. Tsvetanova, D. Klass, J. Salzman and P.O. Brown (2010). “Proteome-Wide Search Reveals Unexpected RNA-Binding Proteins in Saccharomyces Cerevisiae”, PLOS One.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0012671
M. Rabinowitz, D.S. Johnson, J. Salzman, M. Banjevic, C. Cinnioglu, B. Behr (2008). “Reliable Concurrent Calling of Multiple Genetic Alleles and 24-Chromosome Ploidy Without Embryo Freezing Using Parental Support Technology (PS)”, Fertility and Sterility.
http://www.fertstert.org/article/S0015-0282(08)01600-2/pdf
A. Nayak and J. Salzman (2006). “Limits on the Ability of Quantum States to Convey Classical Messages”, Journal of the ACM
https://dl-acm-org.stanford.idm.oclc.org/citation.cfm?id=1120587&dl=ACM&coll=DL
P. Diaconis and J. Salzman (2006). “Projection Pursuit for Discrete Data”, Festschrift for David Freedman, IMS Lecture Notes.
http://projecteuclid.org/download/pdf_1/euclid.imsc/1207580088
M. Jablonski, J. Salzman, A. Wierman and A. P. Godbole (2004). “An Improved Upper Bound for the Pebbling Threshold of the n-path”, Discrete Mathematics.
https://www.sciencedirect.com/science/article/pii/S0012365X03004679
Refereed Conference Proceedings
Ashwin Nayak and Julia Salzman (2002). “On Communication Over an Entanglement-Assisted Quantum Channel”., 17th Annual IEEE Conference on Computational Complexity.
http://ieeexplore.ieee.org/document/1004331
Theses
Barrett, Steven P (2018) “Mechanistic Analysis of Circular RNA Biogenesis from Yeast to Man.” Department of Biochemistry, Stanford University. https://www.proquest.com/docview/2458784423?pq-origsite=gscholar&fromopenview=true
Szabo, Linda (2016) “Precise Detection of Circular and Linear RNA with RNA-Seq.” Department of Biochemistry, Stanford University. https://www.proquest.com/docview/2454712141/E8034D0D745B424APQ/1?accountid=14026
J. Salzman (2007). “Spectral Analysis with Markov Chains." Ph. D. Dissertation, Department of Statistics, Stanford University. http://www.stat.columbia.edu/~jsalzman/thesis.pdf
J. Salzman (2002). “Distinct knots with the same Jones Polynomial.” Princeton University Underdergraduate Thesis.